Slice Simplex (SliceSimplex)¶
Implementation of the slice simplex sampler as described by Cowles et al. [17] for simulating autocorrelated draws of parameters on the simplex 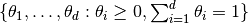 and from a distribution that can be specified up to a constant of proportionality.
and from a distribution that can be specified up to a constant of proportionality.
Model-Based Constructor¶
-
SliceSimplex(params::ElementOrVector{Symbol}; scale::Real=1.0)¶ Construct a
Samplerobject for which slice simplex sampling is to be applied separately to each of the supplied parameters. Parameters are assumed to be continuous and constrained to a simplex.Arguments
params: stochastic node(s) to be updated with the sampler.scale: value0 < scale <= 1by which to scale the standard simplex to define an initial space from which to simulate values.
Value
Returns aSampler{SliceSimplexTune}type object.Example
Stand-Alone Function¶
-
slicesimplex!(v::SliceSimplexVariate, logf::Function; scale::Real=1.0)¶ Simulate one draw from a target distribution using a slice simplex sampler. Parameters are assumed to be continuous and constrained to a simplex.
Arguments
v: current state of parameters to be simulated.scale: value0 < scale <= 1by which to scale the standard simplex to define an initial space from which to simulate values.logf: function that takes a singleDenseVectorargument of parameter values at which to compute the log-transformed density (up to a normalizing constant).
Value
Returnsvupdated with simulated values and associated tuning parameters.Example
################################################################################ ## Multinomial Model ## y ~ Multinomial(n, rho) ## rho ~ Dirichlet(1, ..., 1) ################################################################################ using Mamba ## Data n, k = 100, 5 rho0 = rand(Dirichlet(ones(k))) y = rand(Multinomial(n, rho0)) ## Log-transformed Posterior(rho) + Constant logf = function(rho::DenseVector) logpdf(Multinomial(n, rho), y) end ## MCMC Simulation with Slice Simplex Sampling t = 10000 sim = Chains(t, k, names = map(i -> "rho[$i]", 1:k)) rho = SliceSimplexVariate(fill(1 / k, k)) for i in 1:t slicesimplex!(rho, logf) sim[i, :, 1] = rho end describe(sim) p = plot(sim) draw(p, filename = "slicesimplexplot")
SliceSimplexVariate Type¶
Declaration¶
typealias SliceSimplexVariate SamplerVariate{SliceSimplexTune}
Fields¶
value::Vector{Float64}: simulated values.tune::SliceSimplexTune: tuning parameters for the sampling algorithm.
Constructors¶
-
SliceSimplexVariate(x::AbstractVector{T<:Real})¶ -
SliceSimplexVariate(x::AbstractVector{T<:Real}, tune::SliceSimplexTune) Construct a
SliceSimplexVariateobject that stores simulated values and tuning parameters for slice simplex sampling.Arguments
x: simulated values.tune: tuning parameters for the sampling algorithm. If not supplied, parameters are set to their defaults.
Value
Returns aSliceSimplexVariatetype object with fields set to the values supplied to argumentsxandtune.