Equiv: Bioequivalence in a Cross-Over Trial¶
An example from OpenBUGS [44] and Gelfand et al. [29] concerning a two-treatment, cross-over trial with 10 subjects.
Model¶
Treatment responses are modelled as
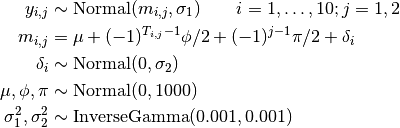
where 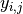 is the response for patient
is the response for patient 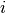 in period
in period 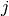 ; and
; and 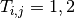 is the treatment received.
is the treatment received.
Analysis Program¶
using Mamba
## Data
equiv = Dict{Symbol, Any}(
:group => [1, 1, 2, 2, 2, 1, 1, 1, 2, 2],
:y =>
[1.40 1.65
1.64 1.57
1.44 1.58
1.36 1.68
1.65 1.69
1.08 1.31
1.09 1.43
1.25 1.44
1.25 1.39
1.30 1.52]
)
equiv[:N] = size(equiv[:y], 1)
equiv[:P] = size(equiv[:y], 2)
equiv[:T] = [equiv[:group] 3 - equiv[:group]]
## Model Specification
model = Model(
y = Stochastic(2,
(delta, mu, phi, pi, s2_1, T) ->
begin
sigma = sqrt(s2_1)
UnivariateDistribution[
begin
m = mu + (-1)^(T[i, j] - 1) * phi / 2 + (-1)^(j - 1) * pi / 2 +
delta[i, j]
Normal(m, sigma)
end
for i in 1:10, j in 1:2
]
end,
false
),
delta = Stochastic(2,
s2_2 -> Normal(0, sqrt(s2_2)),
false
),
mu = Stochastic(
() -> Normal(0, 1000)
),
phi = Stochastic(
() -> Normal(0, 1000)
),
theta = Logical(
phi -> exp(phi)
),
pi = Stochastic(
() -> Normal(0, 1000)
),
s2_1 = Stochastic(
() -> InverseGamma(0.001, 0.001)
),
s2_2 = Stochastic(
() -> InverseGamma(0.001, 0.001)
),
equiv = Logical(
theta -> Int(0.8 < theta < 1.2)
)
)
## Initial Values
inits = [
Dict(:y => equiv[:y], :delta => zeros(10, 2), :mu => 0, :phi => 0,
:pi => 0, :s2_1 => 1, :s2_2 => 1),
Dict(:y => equiv[:y], :delta => zeros(10, 2), :mu => 10, :phi => 10,
:pi => 10, :s2_1 => 10, :s2_2 => 10)
]
## Sampling Scheme
scheme = [NUTS(:delta),
Slice([:mu, :phi, :pi], 1.0),
Slice([:s2_1, :s2_2], 1.0, Univariate)]
setsamplers!(model, scheme)
## MCMC Simulations
sim = mcmc(model, equiv, inits, 12500, burnin=2500, thin=2, chains=2)
describe(sim)
Results¶
Iterations = 2502:12500
Thinning interval = 2
Chains = 1,2
Samples per chain = 5000
Empirical Posterior Estimates:
Mean SD Naive SE MCSE ESS
s2_2 0.0173121833 0.014549568 0.00014549568 0.0007329722 394.02626
s2_1 0.0184397014 0.013837972 0.00013837972 0.0005689492 591.55873
pi -0.1874240524 0.086420302 0.00086420302 0.0032257037 717.76558
phi -0.0035569545 0.087590520 0.00087590520 0.0035141650 621.25503
theta 1.0002921934 0.088250458 0.00088250458 0.0036227671 593.40761
equiv 0.9751000000 0.155828169 0.00155828169 0.0036666529 1806.14385
mu 1.4387396416 0.042269208 0.00042269208 0.0013735876 946.96847
Quantiles:
2.5% 25.0% 50.0% 75.0% 97.5%
s2_2 0.0016375061 0.0056159514 0.013968228 0.024613730 0.053154674
s2_1 0.0017289114 0.0074958338 0.015849718 0.025963832 0.051967161
pi -0.3579631753 -0.2432161807 -0.187946319 -0.130454165 -0.014910965
phi -0.1723722017 -0.0623573600 -0.005681830 0.053144647 0.172913654
theta 0.8416658455 0.9395470702 0.994334281 1.054582177 1.188763456
equiv 1.0000000000 1.0000000000 1.000000000 1.000000000 1.000000000
mu 1.3552569594 1.4110400018 1.438593809 1.466525521 1.519643109