Equiv: Bioequivalence in a Cross-Over Trial¶
An example from OpenBUGS [38] and Gelfand et al. [25] concerning a two-treatment, cross-over trial with 10 subjects.
Model¶
Treatment responses are modelled as
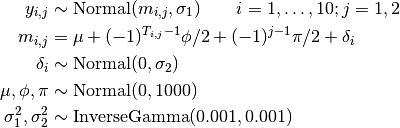
where 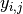 is the response for patient
is the response for patient 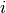 in period
in period 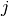 ; and
; and 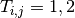 is the treatment received.
is the treatment received.
Analysis Program¶
using Mamba
## Data
equiv = (Symbol => Any)[
:group => [1, 1, 2, 2, 2, 1, 1, 1, 2, 2],
:y =>
[1.40 1.65
1.64 1.57
1.44 1.58
1.36 1.68
1.65 1.69
1.08 1.31
1.09 1.43
1.25 1.44
1.25 1.39
1.30 1.52]
]
equiv[:N] = size(equiv[:y], 1)
equiv[:P] = size(equiv[:y], 2)
equiv[:T] = [equiv[:group] 3 - equiv[:group]]
## Model Specification
model = Model(
y = Stochastic(2,
@modelexpr(delta, mu, phi, pi, s2_1, T,
begin
sigma = sqrt(s2_1)
Distribution[
begin
m = mu + (-1)^(T[i,j]-1) * phi / 2 + (-1)^(j-1) * pi / 2 +
delta[i,j]
Normal(m, sigma)
end
for i in 1:10, j in 1:2
]
end
),
false
),
delta = Stochastic(2,
@modelexpr(s2_2,
Normal(0, sqrt(s2_2))
),
false
),
mu = Stochastic(
:(Normal(0, 1000))
),
phi = Stochastic(
:(Normal(0, 1000))
),
theta = Logical(
@modelexpr(phi,
exp(phi)
)
),
pi = Stochastic(
:(Normal(0, 1000))
),
s2_1 = Stochastic(
:(InverseGamma(0.001, 0.001))
),
s2_2 = Stochastic(
:(InverseGamma(0.001, 0.001))
),
equiv = Logical(
@modelexpr(theta,
int(0.8 < theta < 1.2)
)
)
)
## Initial Values
inits = [
[:y => equiv[:y], :delta => zeros(10, 2), :mu => 0, :phi => 0,
:pi => 0, :s2_1 => 1, :s2_2 => 1],
[:y => equiv[:y], :delta => zeros(10, 2), :mu => 10, :phi => 10,
:pi => 10, :s2_1 => 10, :s2_2 => 10]
]
## Sampling Scheme
scheme = [NUTS([:delta]),
Slice([:mu, :phi, :pi], fill(1.0, 3)),
Slice([:s2_1, :s2_2], ones(2), :univar)]
setsamplers!(model, scheme)
## MCMC Simulations
sim = mcmc(model, equiv, inits, 12500, burnin=2500, thin=2, chains=2)
describe(sim)
Results¶
Iterations = 2502:12500
Thinning interval = 2
Chains = 1,2
Samples per chain = 5000
Empirical Posterior Estimates:
Mean SD Naive SE MCSE ESS
pi -0.1788624 0.07963600 0.0007963600 0.0022537740 1248.5276
mu 1.4432301 0.04735444 0.0004735444 0.0020359933 540.9645
equiv 0.9835000 0.12739456 0.0012739456 0.0022978910 3073.5685
s2_2 0.0187006 0.01466228 0.0001466228 0.0004799037 933.4580
s2_1 0.0164033 0.01412396 0.0001412396 0.0005079176 773.2610
theta 0.9837569 0.07936595 0.0007936595 0.0025041274 1004.5132
phi -0.0195929 0.08005300 0.0008005300 0.0025432838 990.7533
Quantiles:
2.5% 25.0% 50.0% 75.0% 97.5%
pi -0.332750 -0.2265245 -0.184088 -0.1285565 -0.0142326
mu 1.353882 1.4111562 1.441424 1.4740774 1.5345210
equiv 1.000000 1.0000000 1.000000 1.0000000 1.0000000
s2_2 0.001568 0.0061461 0.016302 0.0273322 0.0530448
s2_1 0.001035 0.0043785 0.013677 0.0242892 0.0507633
theta 0.838904 0.9336518 0.974953 1.0320046 1.1567524
phi -0.175659 -0.0686517 -0.025366 0.0315031 0.1456164