Jaws: Repeated Measures Analysis of Variance¶
An example from OpenBUGS [38] and Elston and Grizzle [20] concerning jaw bone heights measured repeatedly in a cohort of 20 boys at ages 8, 8.5, 9, and 9.5 years.
Model¶
Bone heights are modelled as
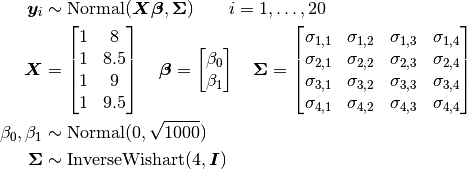
where 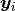 is a vector of the four repeated measurements for boy
is a vector of the four repeated measurements for boy 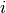 . In the model specification below, the bone heights are arranged into a 1-dimensional vector on which a Block-Diagonal Multivariate Normal Distribution is specified. Furthermore, since
. In the model specification below, the bone heights are arranged into a 1-dimensional vector on which a Block-Diagonal Multivariate Normal Distribution is specified. Furthermore, since 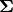 is a covariance matrix, it is symmetric with
is a covariance matrix, it is symmetric with M * (M + 1) / 2 unique (upper or lower triangular) parameters, where M is the matrix dimension. Consequently, that is the number of parameters to account for when defining samplers for 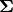 ; e.g.,
; e.g., AMWG([:Sigma], fill(0.1, int(M * (M + 1) / 2))).
Analysis Program¶
using Mamba
## Data
jaws = (Symbol => Any)[
:Y =>
[47.8 48.8 49.0 49.7
46.4 47.3 47.7 48.4
46.3 46.8 47.8 48.5
45.1 45.3 46.1 47.2
47.6 48.5 48.9 49.3
52.5 53.2 53.3 53.7
51.2 53.0 54.3 54.5
49.8 50.0 50.3 52.7
48.1 50.8 52.3 54.4
45.0 47.0 47.3 48.3
51.2 51.4 51.6 51.9
48.5 49.2 53.0 55.5
52.1 52.8 53.7 55.0
48.2 48.9 49.3 49.8
49.6 50.4 51.2 51.8
50.7 51.7 52.7 53.3
47.2 47.7 48.4 49.5
53.3 54.6 55.1 55.3
46.2 47.5 48.1 48.4
46.3 47.6 51.3 51.8],
:age => [8.0, 8.5, 9.0, 9.5]
]
M = jaws[:M] = size(jaws[:Y], 2)
N = jaws[:N] = size(jaws[:Y], 1)
jaws[:y] = vec(jaws[:Y]')
jaws[:x] = kron(ones(jaws[:N]), jaws[:age])
## Model Specification
model = Model(
y = Stochastic(1,
@modelexpr(beta0, beta1, x, Sigma,
BDiagNormal(beta0 + beta1 * x, Sigma)
),
false
),
beta0 = Stochastic(
:(Normal(0, sqrt(1000)))
),
beta1 = Stochastic(
:(Normal(0, sqrt(1000)))
),
Sigma = Stochastic(2,
@modelexpr(M,
InverseWishart(4.0, eye(M))
)
)
)
## Initial Values
inits = [
[:y => jaws[:y], :beta0 => 40, :beta1 => 1, :Sigma => eye(M)],
[:y => jaws[:y], :beta0 => 10, :beta1 => 10, :Sigma => eye(M)]
]
## Sampling Scheme
scheme = [Slice([:beta0, :beta1], [10, 1]),
AMWG([:Sigma], fill(0.1, int(M * (M + 1) / 2)))]
setsamplers!(model, scheme)
## MCMC Simulations
sim = mcmc(model, jaws, inits, 10000, burnin=2500, thin=2, chains=2)
describe(sim)
Results¶
Iterations = 2502:10000
Thinning interval = 2
Chains = 1,2
Samples per chain = 3750
Empirical Posterior Estimates:
Mean SD Naive SE MCSE ESS
beta0 33.64206 1.9166361 0.022131407 0.06737008 809.36637
beta1 1.87509 0.2164271 0.002499085 0.00776451 776.95308
Sigma[1,1] 7.38108 2.7056209 0.031241819 0.18590159 211.82035
Sigma[1,2] 7.19741 2.6922115 0.031086981 0.19045182 199.82422
Sigma[1,3] 6.78381 2.6747486 0.030885337 0.19225335 193.56111
Sigma[1,4] 6.53015 2.6851416 0.031005345 0.19272356 194.11754
Sigma[2,2] 7.53852 2.7705502 0.031991558 0.19576958 200.28188
Sigma[2,3] 7.20718 2.7647599 0.031924698 0.19814088 194.70032
Sigma[2,4] 6.96772 2.7775563 0.032072457 0.19836708 196.05889
Sigma[3,3] 8.03805 2.9649386 0.034236162 0.20539431 208.37930
Sigma[3,4] 8.00528 3.0132157 0.034793618 0.20660383 212.70794
Sigma[4,4] 8.58703 3.1870478 0.036800859 0.21045921 229.31967
Quantiles:
2.5% 25.0% 50.0% 75.0% 97.5%
beta0 29.82471 32.408405 33.654913 34.908169 37.422755
beta1 1.45350 1.726719 1.873572 2.015534 2.307439
Sigma[1,1] 3.87079 5.403112 6.721379 8.779604 14.320056
Sigma[1,2] 3.69391 5.274856 6.515303 8.567864 14.053785
Sigma[1,3] 3.31306 4.888463 6.137073 8.156816 13.266370
Sigma[1,4] 3.02151 4.636266 5.913888 7.860503 13.159120
Sigma[2,2] 3.92078 5.576799 6.857077 8.950953 14.535064
Sigma[2,3] 3.61053 5.233882 6.521158 8.641108 14.040245
Sigma[2,4] 3.29096 4.997585 6.297002 8.382338 13.679823
Sigma[3,3] 4.16453 5.904915 7.321269 9.615860 15.037631
Sigma[3,4] 4.04242 5.850981 7.295268 9.625204 15.275975
Sigma[4,4] 4.37143 6.340437 7.843085 10.166456 16.465667