Salm: Extra-Poisson Variation in a Dose-Response Study¶
An example from OpenBUGS [38] and Breslow [8] concerning mutagenicity assay data on salmonella in three plates exposed to six doses of quinoline.
Model¶
Number of revertant colonies of salmonella are modelled as
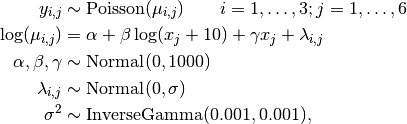
where 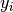 is the number of colonies in plate
is the number of colonies in plate 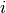 and dose
and dose 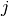 .
.
Analysis Program¶
using Mamba
## Data
salm = (Symbol => Any)[
:y => reshape(
[15, 21, 29, 16, 18, 21, 16, 26, 33, 27, 41, 60, 33, 38, 41, 20, 27, 42],
3, 6),
:x => [0, 10, 33, 100, 333, 1000],
:plate => 3,
:dose => 6
]
## Model Specification
model = Model(
y = Stochastic(2,
@modelexpr(alpha, beta, gamma, x, lambda,
Distribution[
begin
mu = exp(alpha + beta * log(x[j] + 10) + gamma * x[j] + lambda[i,j])
Poisson(mu)
end
for i in 1:3, j in 1:6
]
),
false
),
alpha = Stochastic(
:(Normal(0, 1000))
),
beta = Stochastic(
:(Normal(0, 1000))
),
gamma = Stochastic(
:(Normal(0, 1000))
),
lambda = Stochastic(2,
@modelexpr(s2,
Normal(0, sqrt(s2))
),
false
),
s2 = Stochastic(
:(InverseGamma(0.001, 0.001))
)
)
## Initial Values
inits = [
[:y => salm[:y], :alpha => 0, :beta => 0, :gamma => 0, :s2 => 10,
:lambda => zeros(3, 6)],
[:y => salm[:y], :alpha => 1, :beta => 1, :gamma => 0.01, :s2 => 1,
:lambda => zeros(3, 6)]
]
## Sampling Scheme
scheme = [Slice([:alpha, :beta, :gamma], [1.0, 1.0, 0.1]),
AMWG([:lambda, :s2], fill(0.1, 19))]
setsamplers!(model, scheme)
## MCMC Simulations
sim = mcmc(model, salm, inits, 10000, burnin=2500, thin=2, chains=2)
describe(sim)
Results¶
Iterations = 2502:10000
Thinning interval = 2
Chains = 1,2
Samples per chain = 3750
Empirical Posterior Estimates:
Mean SD Naive SE MCSE ESS
gamma -0.0011251 0.00034537 0.000003988 0.00002158 256.1352
alpha 2.0100584 0.26156943 0.003020344 0.02106994 154.1158
s2 0.0690770 0.04304237 0.000497010 0.00192980 497.4697
beta 0.3543443 0.07160779 0.000826856 0.00564423 160.9577
Quantiles:
2.5% 25.0% 50.0% 75.0% 97.5%
gamma -0.0017930 -0.0013474 -0.001133 -0.0009096 -0.0003927
alpha 1.5060295 1.8471115 2.006273 2.1655604 2.5054785
s2 0.0135820 0.0397881 0.059831 0.0874088 0.1774730
beta 0.2073238 0.3127477 0.358298 0.4000692 0.4878904